Contact Us
Latest News
- Clues beginning to emerge on asymtomatic SARS-CoV-2 infection
- Back in November of 2020, during the first wave of the COVID-19 pandemic, I was teaching an in-person microbiology laboratory. One of my students had just been home to see his parents, and they all c…
- Read more
- Could there maybe be better uses of genetics and probiotics?
- Professor Meng Dong and his laboratory have created a probiotic that can metabolize alcohol quickly and maybe prevent some of the adverse effects of alcohol consumption. The scientists cloned a highl…
- Read more
- ChatGPT is not the end of essays in education
- The takeover of AI is upon us! AI can now take all our jobs, is the click-bait premise you hear from the news. While I cannot predict the future, I am dubious that AI will play such a dubious role in…
- Read more
- Fighting infections with infections
- Multi-drug-resistant bacterial infections are becoming more of an issue, with 1.2 million people dying of previously treatable bacterial infections. Scientists are frantically searching for new metho…
- Read more
- A tale of two colleges
- COVID-19 at the University of Wisconsin this fall has been pretty much a non-issue. While we are wearing masks, full in-person teaching is happening on campus. Bars, restaurants, and all other busine…
- Read more
Chapter 2 - Fundamental concepts for understanding microbiology
2 - 1 Introduction
Microbiology stands on the shoulders of chemistry, physics, mathematics, and many other areas of study. To be able to talk the language of microbiology, there is an entrance fee in the form of a basic set of knowledge you must acquire from these other disciplines. In this chapter, we cover many important concepts that a student of microbiology needs as a foundation for the rest of microbiology. These bases concepts are probably a review for many, but you may want to read it anyway because it might give you a new perspective on the knowledge you already have.
2 - 2 Fundamental Properties of Matter
Learning Objectives
After reading this section, students will be able to...
- Explain the composition of matter.
- Describe the atomic structure of the common elements in biological items.
- Explain the forces that hold molecules together, both covalent and non-covalent.
Everything we see, everything we touch and taste, most everything in our natural world is composed of matter. Matter surrounds us and interacts with us, but what is it really? Matter is anything that occupies space and has mass. Each bit of matter has a characteristic density, hardness, taste and color. Matter can undergo transformations including melting, boiling, and changes in composition. But, what is it made of and how does that determine the above properties? To begin, consider this analogy.
The writing on this page can be broken down into its basic parts. Each sentence is composed of words. Each word contains 26 letters of the English alphabet, and each letter is simply a combination of curves, lines, and dots. Building these curves, lines, and dots into words and sentences can convey ideas such as the nature of matter. Similarly, matter is composed of atoms, and each atom can be broken down into subatomic particles. The way these subatomic particles combine determines the properties and behaviors of the atoms and the matter they comprise.
Surprisingly, in this vast cosmos we live in, there are only 109 natural atoms (also called elements). All of them are combinations of three subatomic particles, protons, neutrons, and electrons. Protons are positively charged particles that have a tiny mass. Neutrons are of similar mass but have no charge. Electrons are particles of very little mass with a charge equal to and opposite to that of the proton. It is convenient when talking about such a small charge and mass to use relative units, and the weight of one proton is defined as one atomic unit (also be referred to as a Dalton). For a similar reason, the charge on a proton is positive and given the value of +1. The charge on an electron is negative and given the value -1.
Table 2-1. Properties of subatomic particles
| Subatomic Particle | Actual Charge (Coulomb) |
Relative Charge | Actual Mass (g) | Relative Mass | Mass in atomic Units or Daltons |
| Electron | 1.6 x 10 -19 | -1 | 9.11 x 10 -28 | 1/1837 | 0 |
| Proton | 1.6 x 10 -19 | +1 | 1.67 x 10 -24 | 1 | 1 |
| Neutron | 0 | 0 | 1.67 x 10 -24 | 1 | 1 |
Protons and neutrons combine to form a nucleus in the center of the atom. The positive charge of the nucleus attracts and holds electrons. The electrons move very fast (near the speed of light) and, as best as we can measure, form a cloud of charge around the nucleus. Electrons want to be around as much positive charge as possible, and protons want to collect as much negative charge as possible. The ideal state for the atom, its most stable state, is to balance these two forces such that the atom has an equal amount of positive and negative charge. As an example, here is the construction of two of the lighter elements, hydrogen and helium. Hydrogen has one proton (one positive charge), no neutrons, and one electron (one negative charge). Helium has two protons, two neutrons, and two electrons (Figure 2.1).
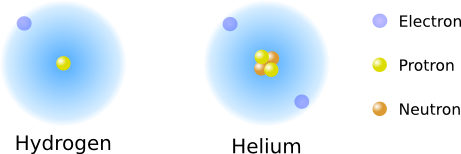
Figure 2.1. Arrangement of Hydrogen and Helium.. The arrangement of protons, neutrons and electrons in hydrogen and helium
The number of protons in an atom increases its weight and changes its fundamental properties, differentiating one element from the next. The number of protons in an atom's nucleus delineates its properties, and scientists coined the term atomic number to express this idea. An atom containing 6 protons is a carbon atom no matter how many neutrons or electrons it contains.
The addition of a neutron adds to an atom's weight and can affect its stability, but it does not change its properties. In fact, atoms of carbon containing seven neutrons (carbon 13) and eight neutrons (carbon 14) exist in nature. Carbon 12, Carbon 13, and Carbon 14 are three isotopes of carbon.
Bonding of atoms
The electron count of an atom affects its reactivity. Atoms want to be "full" of electrons. If they are not, they will try to borrow them from other atoms. As an example, consider hydrogen and helium. Hydrogen is a very reactive molecule, always searching for an extra electron, while helium having two electrons is "full" and not very reactive.
So what determines whether an atom is full or not? The way electrons associate with the nucleus. The discipline of quantum mechanics elucidated many of the rules for this association. When electrons come in association with the nucleus of an atom, they want to house themselves in very precisely defined spaces. These acceptable spaces are called orbitals. Each orbital defines a region where the electron is likely to be found and contains two electrons when full. Atoms have discrete energy levels or shells that contain these orbitals. Electrons must occupy orbitals in these energy states and are not allowed to exist outside the shells. A way you can imagine this is as the difference between sitting on an inclined plane vs. a set of steps. When on a plane, you can have any amount of potential energy, but you can only have discrete amounts with a set of steps. As one moves away from the nucleus, these shells require higher energy electrons to fill the orbitals. Atoms prefer to be in the lowest energy level possible; thus, electrons will fill the open orbitals in the lowest energy shell first, then move to the next shell, and so on.
As an analogy, orbitals can be thought of as houses where two electrons can live. These houses are organized into subdivisions (the shells) encircling a lake (the nucleus) at the bottom of a valley. Paths away from the hill have large steps instead of a simple incline. The most desirable location is on the lakefront, and electrons fill that house first. Further sets of electrons will fill the closest available house to the lake, but they must jump up a step, a quantum of energy to do so. More energy is required to live in the houses further from the lake because you have to walk up the steps to get there (Figure 2.2).
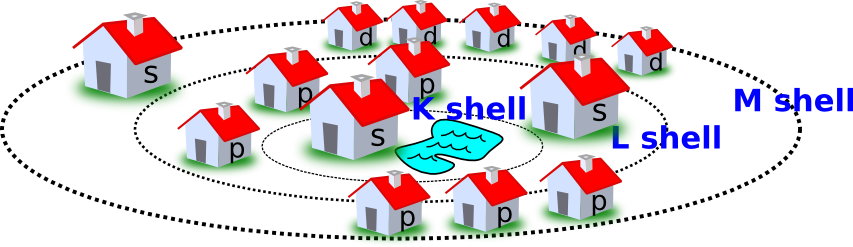
Figure 2.2. The houses of electrons. A useful analogy for thinking about electron positions in atoms
The rules for these associations determine the number and types of orbitals present in each shell. The first shell, the K shell, contains one s orbital that is spherical. The next shell, the L shell, contains four orbitals, one s orbital, and three p orbitals. The M shell contains 1 s, 3 p, and 5 d orbitals. Higher shells will contain more orbitals, but for the most part, atoms important in biological systems do not fill beyond the M shell (Figure 2.3).
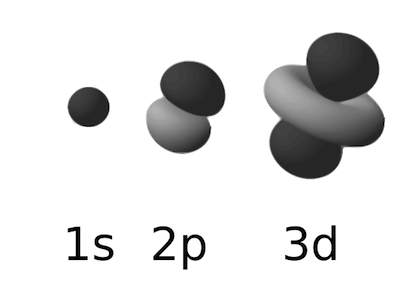
Figure 2.3. Examples of atomic orbitals. The most common types of atomic orbitals in biological systems; the s, p and d orbitals. Figure adapted from a figure in the wikimedia commons. (http://commons.wikimedia.org/wiki/File:Electron_orbitals.svg)
Let's now go back to hydrogen and helium. Hydrogen has one electron that fills the s orbital of the K shell. There is room for one more electron in the orbital, and hydrogen seeks it out. In its elemental form, each hydrogen atom will pair up with a second hydrogen atom, forming H2. Helium contains two electrons that fill the s orbital of the K shell. The K shell is full, and helium does not easily add more electrons. Elements with completely full shells (helium, argon, neon, and krypton, for example) are very non-reactive
In contrast to helium, most atoms have partially filled outer shells and are constantly searching for ways to fill them. This tendency makes them reactive with other atoms, and that is why they associate and build larger structures called molecules. Atoms fill these orbitals by sharing electrons between different nuclei and overlapping their orbitals. The number of protons and electrons in an atom dictates how much sharing each atom is willing to do and dictates its behavior in chemical reactions. Table 2.2 lists some common elements in organisms and their electron-pairing behavior.
Table 2-2. Common elements in living systems and their properties.
|
Element |
Symbol |
Atomic Number |
Atomic Weight |
Protons |
Neutrons |
Electrons |
Electrons in outer shell |
Bonds Formed |
| Hydrogen | H | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| Carbon | C | 6 | 12 | 6 | 6 | 6 | 4 | 4 |
| Nitrogen | N | 7 | 14 | 7 | 7 | 7 | 5 | 3 |
| Oxygen | O | 8 | 16 | 8 | 8 | 8 | 6 | 2 |
| Phosphorus | P | 15 | 31 | 15 | 16 | 15 | 5 | 3 or 5 |
| Sulfur | S | 16 | 32 | 16 | 16 | 16 | 6 | 2, 4 or 6 |
All sorts of combinations between these molecules are possible. Carbon can combine with four hydrogens to form methane. Nitrogen can combine with another nitrogen molecule to form nitrogen gas. Pure oxygen can share electrons with another oxygen atom, forming O2. Two hydrogens can share their electrons with an oxygen atom forming water. Figure 2.4 shows some simple molecules.
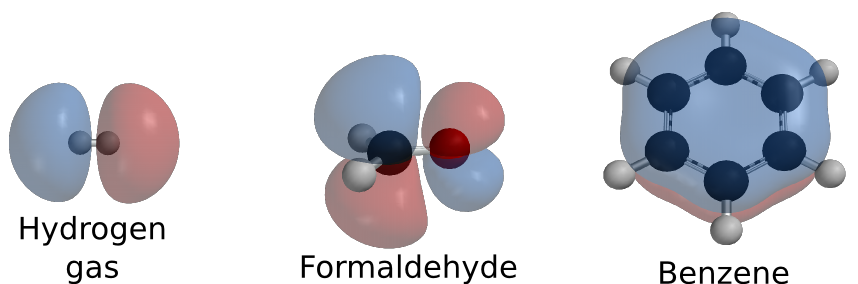
Figure 2.4. Some simple molecules. A few examples of molecules that can be formed. These molecules are drawn showing their orbitals. Orbital drawings created by Ben Mills and available from wikimedia commons
The term covalent bond is used to describe this type of electron sharing. Covalent bonds are stable and bind molecules together rather tightly. You can view this electron sharing as a combination of the individual shared electrons forming a distributed electron cloud around the two molecules.
Covalent bonds are not the only ways to interact
As we have learned above, covalent bonds connect atoms forming the monomers and polymers of the cell, but there are other forces at work. Three of the more essential bonding forces are ionic interactions, hydrogen bonds, and hydrophobic interactions, as shown in Figure 2.5.
Ionic interactions are the attraction of opposite charges for one another. Negatively charged groups are attracted to positively charged groups.
Hydrogen bonds involve hydrogen, which is not very good at holding onto electrons when covalently bonded with certain other atoms (usually oxygen or nitrogen). It will not get its fair share of time with the electrons and will be attracted to high electron densities around other nearby atoms. The hydrogen atoms are attracted to the extra electrons and tend to stay in the vicinity of the electron density. Not surprisingly, oxygen and nitrogen atoms have these high electron densities. This is not a covalent bond and hydrogen bonds tend to be weak interactions, but large numbers of them can add significantly to the stability of a protein or a DNA double helix.
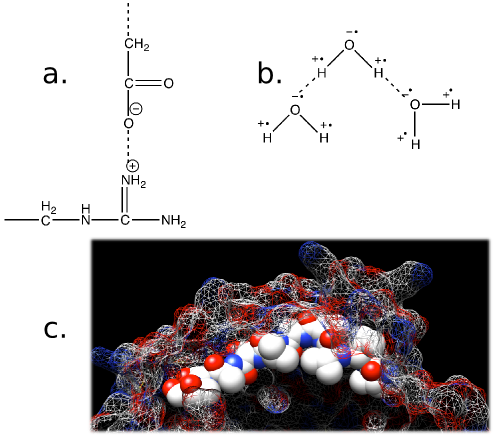
Figure 2.5. Forces that affect biological systems. Show in the figure are a) Ionic interactions. The nitrogen (N) on the side chain of arginine is positively charged and is attracted to the negative charge on the carboxyl group. b) Hydrogen bonds. The hydrogen atoms in water are starved for electrons and the oxygen is electron rich. This will cause the hydrogen atoms to be attracted to the electron dense oxygen. c) Hydrophobic interactions. Non-polar molecules, such as some side chains on amino acids, will disrupt the hydrogen bonding of water and make the system energetically unfavorable. Highlighted is a string of amino acids that are hydrophobic (Val Val Val Thr Ala Val Thr Ala Thr Thr) in the KasA protein. Notice how all the hydrophobic groups, the white spheres, are buried under the surface of the protein (shown as a mesh). The KasA protein is a protein made by Mycobacterium tuberculosis and is part of the pathway for synthesizing long-chain fatty acids. These are important for its virulence and is a target for new drugs.
If you want to see hydrogen bonds in action, fill a glass of water to overflowing. Note how the water clings to itself, even though it is above the height of the glass. Hydrogen bonds between the water molecules cause them to stick together. Now add a few drops of detergent. The detergent will disrupt the bonding between the water molecules. Don't do this unless you have a towel nearby!
Hydrophobic interactions consist of the congregation of non-polar groups together. The presence of a hydrophobic (water-hating) molecule in a water solution causes an undesirable ordering of the water around it. Water hates this and will minimize this interaction by pushing the hydrophobic groups together and away from the water. Think back to the behavior of oil in water. When you place some oil in a bottle of water, the oil tends to separate from the water and form one large bleb (that is a scientific term, really, look it up). If you shake the bottle, the oil disperses, but in a little while, it all congregates again. This behavior is hydrophobic interaction at work. The same process is functioning at the molecular level. Hydrophobic groups in biological systems consist of alkyl chains (see later in the chapter for a definition of this term). These groups are pushed together and hide away from the water in the environment. This grouping can lead to the formation of large structures spontaneously. Hydrophobic interactions largely drive membrane formation and protein folding.
Key Takeaways
- Matter is composed of atoms, and atoms are made of protons, neutrons and electrons.
- The properties of an atom are dictated by the number of protons in its nucleus.
- Electrons are housed in shells of discrete energy, and each shell will have various orbitals. Each orbital can house 2 electrons.
- Most atoms have partially filled shells and will seek out other atoms to pair with, forming covalent bonds.
- Living systems are mostly composed of the lighter elements of the periodic table, H, C, N, O, P, and S.
- Covalent bonds are not the only way for molecules to interact. Forces such as hydrophobic interactions, ionic interactions, and hydrogen bonds are important in biological systems.
2 - 3 How we describe simple molecules
Learning Objectives
After reading this section, students will be able to...
- Be able to read and write the chemical and structure formula of important molecules.
- Be able to recognize common carbon groups found in biological molecules.
- Describe the properties of water that make it so crucial for biological life.
- Define pH and identify its importance in living systems.
- Explain the unique properties of carbon and how these make it central in biological structures.
A common terminology for representing atoms and compounds has been developed to help communicate their composition and the forces that hold them together. The most important for our purpose are the chemical formula and the structural formula. The chemical formula is a type of shorthand that lists the makeup of any compound. Table 2.2 lists the symbols used for the common elements in biological molecules. A chemical formula contains the symbol for each constituent atom and its ratio in the compound. If more than one atom of the same type is present in a molecule, a subscript following the symbol designates the number (Table 2.3). The structural formula shows the arrangement of atoms in the compound. Each atom of the molecule is drawn using its symbol, and lines are drawn between them, indicating the covalent bonds formed.
Table 2.3. Comparison of Chemical Formula, Structural Formula, and Molecular Weight.
| Compound | Chemical Formula | Structural Formula§ | Molecular Weight* | Comments |
| Hydrogen | H2 | 1 | 2.01 | |
| Methane | CH4 | 2 | 16.04 | The major component of natural gas |
| Ethanol | C2 H6 O | 3 | 46.06 | Common alcohol found in beer, wine and liquors |
| Acetic acid | C2 H4 O2 | 4 | 60.05 | Major component of vinegar |
| Glucose | C6 H1 2 O6 | 5 | 180.16 | Common sugar in candies and soft drinks |
| Sucrose | C1 2 H2 2 O1 1 | 6 | 342.30 | Table sugar |
| Oleic Acid | C1 8 H2 4 O2 | 7 | 282.47 | Major fatty acid in butter |
§ Structural formulas are shown in Figure 2.6
*Molecular weights are never exact integers dues to the existence of isotopes for each element. The atomic weight of each atom is an average of the isotopes commonly found.
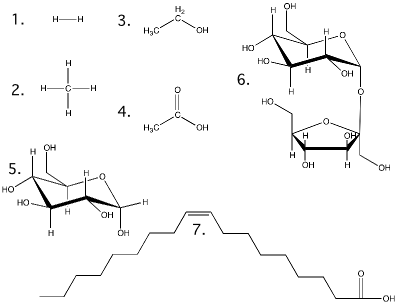
Figure 2.6. Structural formulas for the molecules in Table 2.3. Show here are the structures for the molecules discussed in table 2.3
In biological chemistry, much of the structure of a molecule is made up of carbon and hydrogen atoms. Carbon will form the molecule's backbone, and hydrogen will take up extra bonds that do not link to any other atom. Organic chemists have developed a further shorthand of not writing in the hydrogen atoms and leaving out the carbon symbol. This shorthand greatly reduces the work of drawing structures and increases the structural clarity of the molecule. We will follow this convention throughout most of this book.
It is also important to understand the procedure for measuring and weighing atoms and molecules. The weight of an atom is the sum of the protons and neutrons it contains in its nucleus. The molecular weight is the sum of the weights of all the atoms that make up the molecule. Since it is difficult to work with one molecule of anything, scientists created the term gram molecular weight. The gram molecular weight is the weight of 6.022 x 1023 molecules of a compound. For each molecule, this works out to exactly the molecular weight of the compound in grams. For example, the molecular weight of water is 18 Daltons (1 oxygen : 16 Daltons; 2 hydrogen: 2 Daltons), and the gram molecular weight is 18 grams. A mole of a compound is defined as one gram molecular weight. Finally, the concentration of a compound dissolved in water is most conveniently expressed as molarity. Scientists define this as the number of moles of a compound per liter of water. Figure 2.7 shows a summary of these conversions.
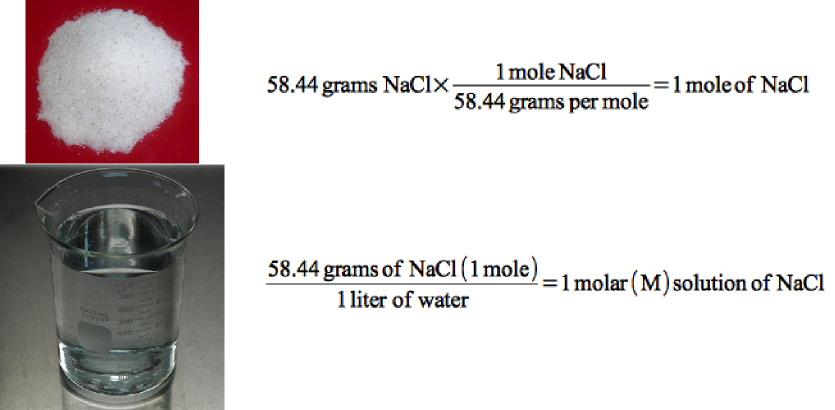
Figure 2.7. Conversion of grams to moles to molarity.. Sodium Chloride has a molecular weight of 58.44, which is the weight of 6.022 x 1023 molecules. If 58.44 grams are weight out, that would be one mole of NaCl. If this is then dissolved in water it would be a 1 molar solution of NaCl.
Water
Water is the most common molecule on the earth's outer layer, covering 75% of the earth's surface. Because of its unique properties, it is the universal solvent for the chemistry of life. You are more than 60% water and would die if your supply were cut off for more than a few days. The oxygen atoms in water pull the electrons toward themselves, causing the hydrogen atoms to be electron starved. The hydrogen in water will seek to form hydrogen bonds with the oxygen of nearby water molecules. This stickiness is termed polarity. Water is not the only polar molecule, any molecule where the electrons are not shared equally will be polar, but water is one of the smallest and most common polar molecules. Because of its polarity and size, water has several unique properties that make it so essential to living systems:
- Water is an excellent solvent. More things dissolve in water than any other solvent. Because of its polar nature, water can cause ionic compounds, like salt, to dissociate into their constituent atoms freeing them to participate in chemical reactions. Enzymes and many of the biomolecules of living systems are soluble in water.
- Water sticks to things, which results in capillarity, the ability to move through small orifices or fine pores against the force of gravity and other forces. This tendency is crucial for moving water about in plants, animals, and microorganisms.
- Water has a high specific heat, meaning that it requires a large amount of heat to change its temperature. Since organisms are mostly water, they lose and gain heat slowly, providing a stable or slowly changing temperature.
- Because of its polarity, water has a high boiling point allowing life to exist in a large range of temperatures.
pH profoundly affects biological activities
The cell cytoplasm is an aqueous environment and acids can dissociate into hydrogen ions (H+) and anions. The higher the H+ concentration, the more acidic the solution. Bases in solution can also dissociate into OH- ions and cations. A more basic solution will have a higher concentration of OH-. This balance in a solution can be quantified by measuring the hydrogen ion concentration, which can vary over a wide range. The concept of pH (potential of hydrogen) was developed as a kind of shorthand for the concentration of hydrogen ions and is defined in the following equation
pH = -log[H+]
Where H+ is the concentration of hydrogen ions in moles per liter.
In normal aqueous solutions, pH values will range from 0 to 14. A pH of 14 indicates a hydrogen ion concentration of 1 x 10-14 moles per liter. A pH of 0 indicates a hydrogen ion concentration of 1 mole per liter, a very acidic solution! Human body fluids have a pH of about 7 or a neutral pH where the concentration of hydrogen ions equals the concentration of hydroxide ions. Acidic solutions contain more H+ than OH- and have a pH below 7, while basic solutions have more OH- than H+ and have a pH above 7. Figure 2.8 shows the pH scale and the pH of some fluids you are probably familiar with.
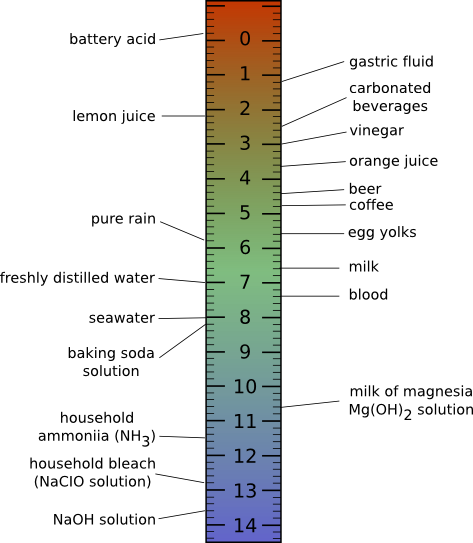
Figure 2.8. The pH scale and the pH of some common foods.. At low pH, the hydrogen ion concentration is high, as the pH increases, the concentration of hydrogen ions decrease and the concentration of hydroxide ions increase. Acidic solutions have more hydrogen ions and basic solutions have more hydroxide ions. The pH of common items is also shown
Maintaining the pH of the cytoplasm is critical to cell function. Small changes in pH can have profound effects on the cell. Many enzymatic reactions require the use of H+ or OH- and changing the pH of the cytoplasm can cause them to no longer function. In addition, enzyme stability can be affected by pH, causing enzymes to denature. Larger changes can even cause the cells chromosome to destabilize.
The cells collection of acids and bases is constantly changing as it moves through its environment, takes up nutrients, carries out metabolism, and excretes waste products. Cells maintain their pH using molecules that can serve as buffers. Buffers are molecular sponges that soak up H+ or OH-. These molecules readily react with H+ and OH- and prevent drastic changes to the pH of the cytoplasm. Microbes that grow in extreme environments of very high or low pH will even have pumps that pump H+ and/or OH- in or out of the cell. The goal is always to maintain the cytoplasm at a near-neutral pH. In low pH environments, cell pump H+ out, while in high pH environments, cells cell pump H+ in.
In contrast to the cytoplasm of the cell, the outside environment can have a wide range of pH, which can dictate what types of microbes can thrive. Soils and water, often near-neutral pH, can become acidic or basic depending upon the mix of minerals, pollutants, and the action of microbes themselves. Thiobacillus ferroxidans can grow in high sulfur coal piles, converting the sulfur to sulfuring acid. If industries leave coal piles in the open air, water flowing through them can become rivers of sulfuric acid, as low as pH 2. This microbe will happily grow under these conditions. Microbes growing in a laboratory medium, or in milk, can produce acidic end products that drop the pH. This result is rarely desirable in the laboratory, and buffers are often added to media to prevent pH changes. However, the growth of microbes in milk, and it's acidification, is often desirable in the formation of delectable treats such as yogurt, buttermilk, and cheese. No matter the pH of the environment, there are almost always microbes capable of growing under these conditions.
Carbon is particularly central to biology
Carbon can rearrange its 1s and 3 p orbitals in its outermost unfilled shell into four equal orbitals that project out in a pyramid-like organization. Each orbital contains one electron, forming four equal bonds instead of the two one would typically expect. This unique property enables it to bond with other atoms to form intricate lattices of infinite variation. Organic molecules (as carbon-containing compounds are called) use carbon backbones as the skeleton for building all sorts of structures that can perform seemingly limitless functions. If carbon did not have this odd behavior, life would be very different (or maybe not even possible). Figure 2.9 shows bonding in carbon atoms.
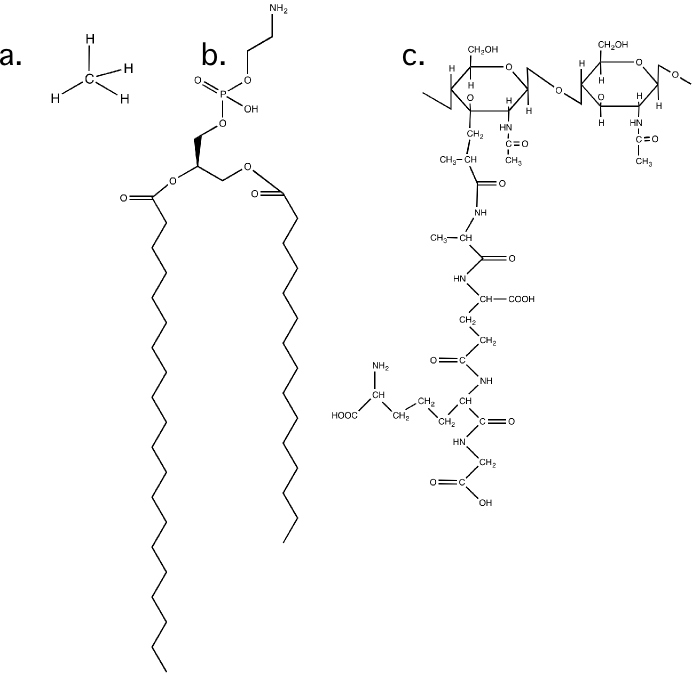
Figure 2.9. The arrangement of bonds in a carbon molecule. a) The structure of methane. Note how each bond is equidistant from the others b) A larger carbon molecule, in this case a membrane lipid. c) The structure of of a peptidoglycan molecule. These units will be put together to form part of the bacterial cell wall.
The names of carbon groups
As we examine the prokaryotes, certain chemical assemblies involving carbon will pop up over and over. Scientists have developed terms for these groups, and it is useful to learn this jargon to facilitate discussions about how the molecules of life behave and are put together. There are only a few of them, and it is worth spending the time to commit them to memory. These structures have important chemistries in biological molecules that help the larger structures in the cell perform their functions.
Table 2-4. Carbon groups often found in organic structures
| Group Name | Group Structure | Comments |
| Alcohol or Hydroxyl | 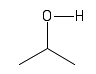 |
Hydroxyl groups decorate many different molecules and are important in chemical reactions and in holding larger molecules together. |
| Keytone | 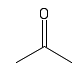 |
Carbon double bonded to oxygen. Important in reactions and structure. Often found in sugars |
| Aldehyde | 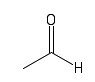 |
A keytone at the end of a molecule. These are also often found in sugars |
| Carboxyl | 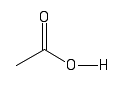 |
Under normal cell conditions, these groups will often lose there hydrogen. They are acidic and have a negative charge. |
| Amino | 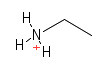 |
Nitrogen containing groups. They are found in proteins, DNA and many other structure in the cell. They will often pick up an extra hydrogen from the environment and give their containing molecule a positive charge. |
| Methyl | 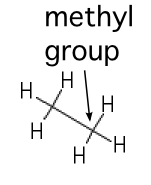 |
A carbon saturated with hydrogens. These groups tend to repel water |
| alkyl | Long chains of carbons tend to repel water and are useful for ordering structures in the cell. |
Stereochemistry is important for biological systems
In biological systems, stereoisomers are significant, especially in the case of sugars and amino acids. In many cases, organisms and their enzymes will only work with one type of molecule and not the other in much the same way as a glove only fits on one hand. Sugars that have identical molecular formulas but are mirror images of one another have been given the designations D and L to distinguish them. Biological systems, for the most part, prefer the D form of sugars. Similarly, amino acids can exist in either the D or L form, but in this case, life prefers the L form of amino acids (except for glycine, which does not have stereoisomers) (Figure 2.10). Why was one form chosen over the other? It was most likely a random event of evolution - when proteins were first evolving, L amino acids happened to be used to construct them, and this bias has been passed down through the ages. There are a few examples of the use of D amino acids for specific functions in living systems, for example, as part of the cell wall of bacteria.

Figure 2.10. The two isomers of alanine.. The L form of alanine is used in proteins, while the D form only finds use in the cell wall of bacteria.
We should note that a more modern system has been developed for naming stereoisomers. This system happens to use R (rectus) and S (sinister) as designations, but more importantly establishes a consistent method for naming any isomeric compound. The details are not important, but the student will see both D/L and R/S forms when referring to these types of geometries. Unfortunately for historical continuity, the D and L designations are used for amino acids and sugars and complicate things a bit when learning.
Key Takeaways
- A chemical formula and a structural formula are two ways to represent molecules. A chemical formula tells you the identity and number of each element, while a structural formula indicates how the elements are arranged in the molecule.
- Measuring and weighing molecules uses a system based upon the natural mass of atoms. Molecule weights are expressed as their molecular weight, or gram molecular weight. The amounts of molecules are expressed in moles, and their concentrations in solutions are expressed as molar.
- Water is the universal solvent for biological systems.
- The concentration of hydrogen ions, the pH, greatly influences biological chemistry and cells work hard to maintain an appropriate pH
- Carbon has the useful ability to rearrange its s and p orbitals, making it capable of forming four bonds instead of two. This allows the chaining together of an infinite number of carbons in unique arrangements, enabling the construction of life.
- Life has a handedness to it. D forms of sugars and L forms of amino acids are mostly used in the construction of biological molecules.
2 - 4 Shorthand for chemical reactions
Learning Objectives
After reading this section, students will be able to...
- Interpret a chemical equation describing the substrates and products of the reaction.
- Define what free energy is and how it relates to chemical reactions.
- Explain an oxidation-reduction reaction and give an example of one.
Just as scientists use a shorthand to represent atoms and molecules in the cell, there is also a shorthand for drawing reactions. Chemical equations diagram the conversion of molecules from one structure to another (Figure 2.11). An equation will show two sets of chemical or structural formulas separated by an arrow indicating the direction of the reaction. Typically, on the left side will be the reactants, and on the right side will be the products. The arrow between the two can point only one way or both ways, indicating a reversible reaction. Most reactions proceed in both forward and reverse directions. If neither the reactants nor products exit a system, the reaction will proceed toward an equilibrium, at which point the forward and reverse reactions will occur at the same rate. Chemical equations must be balanced, containing the same number and types of atoms on each side of the reaction.
As an example of a chemical reaction, consider what happens when you put oxygen gas and hydrogen gas together in a container and light a match; an explosion will occur, forming water. This reaction is reversible. If water is placed in a test tube and an electric current is applied, hydrogen and oxygen gas will bubble up from the solution.
![]()
Figure 2.11. A simple chemical equation. The conversion of hydrogen gas and oxygen to water. Note that the reverse is also possible.
Reactions in the cell are driven by the amount of free energy they produce and by concentration
Organisms take energy and use it to rearrange chemicals with the ultimate goal of producing more of themselves. There are thousands of chemical reactions in a cell. To get each of these needed chemical rearrangements to occur, the total reaction has to be favorable. These favorable reactions can result in free energy that can be captured for later or used to drive the construction of something. No reaction in the cell will take place if it isn't favorable. In food digestion, these reactions are naturally favorable and net the organism extra energy. When the cell is building something, like a protein or cell wall, the reactions by themselves are not favorable. The cell drives these along by linking them with the release of chemical energy, often the breakdown of high-energy phosphate compounds. The breakdown of the phosphate compound in these reactions is dependent upon the building of the desired product. Since more energy is released from the phosphate than is used to make the product, the reaction becomes favorable.
The concentration of substrates and products also has a tremendous influence on the rate of any reaction. A large amount of substrate will increase the rate of a reaction and a large amount of product can inhibit it. This property has two implications for the cell. First, substrates have to be of a high enough concentration so that the reactions can occur. Since compounds in the environment are typically at low concentrations, the cell must accumulate them against that concentration gradient. Second, the cell sometimes pulls reactions along by immediately removing the product. Removal of product is especially effective when reactions are reversible and result in similar concentrations of substrates and products.
Oxidation-reduction reactions as a special case
A common theme in many reactions in the cell is the shuttling of electrons from one molecule to another. The biosynthesis of many cell components and the generation of energy in most microbes involves electrons changing molecular hands. These are termed oxidation-reduction reactions or redox reactions, and many microbiology students have difficulty understanding how they work. As a first attempt, that will oft be repeated to make sure you get the point, we will now expose them at their most intimate level. In a redox reaction, electrons pass from one molecule, which is oxidized, to a second molecule, which is reduced. As an example, lets look at one of the steps of glycolysis, a digestive pathway that you have in your cells. (Figure 2.12)
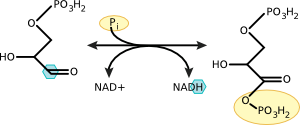
Figure 2.12. Oxidation of glyceraldehyde 3-phosphate to 1,3 bisphophoglycerate. Note the removal of electrons from the carbon molecule and their addition to NAD Electrons are removed from the highlighted carbon and donated to NAD+.
Glyceraldehyde has an aldehyde group associated with its first carbon. This aldehyde carbon is attached to one carbon, one oxygen, and one hydrogen. It shares four electrons, one with carbon, one with hydrogen, and two with oxygen. During the reaction, the hydrogen and its electron are pulled off of glyceraldehyde-3-phosphate and given to NAD. This oxidation leaves carbon with an unfilled orbital, and it reacts with a nearby phosphate to satisfy that need, forming 1,3 bisphosphoglycerate. In this reaction, glyceraldehyde is oxidized, it loses electrons, and NAD is reduced, it gains electrons. We will discuss these ideas in more detail in the chapter on metabolism.
This ends our review of basic chemistry. We now move on to consider these chemical concepts in the context of biological molecules. In this chapter, we will review the common monomers and simple polymers that make up the cell: sugars, nucleic acids, proteins, and lipids. In Chapter 3, we will examine how the cell puts together these building blocks to make bacterial cell structures.
Key Takeaways
- Chemical equations diagram the conversion of reaction substrates into reaction products.
- A reaction is favorable if free energy is released and is available to do work. No reaction in the cell will occur unless it is favorable.
- Oxidation-reduction reactions are a special case of chemical reaction where electrons are moved from one molecule to another.
2 - 5 Sugars are common components of the cell
Learning Objectives
After reading this section, students will be able to...
- Describe why the cell makes polymers and the important polymers of the cell.
- Explain the size and structure of sugars
- Explain how sugars monomers are put together to make polymers.
- Explain the chemical and structural differences between DNA and RNA.
Monomers and Polymers
Living systems are made of organic molecules, and the cell has settled on just a few types of molecules to make up the majority of its structures: sugars, nucleic acids, amino acids and lipids. It is somewhat shocking that the list is so short, but it demonstrates the elegance of the whole system.
Monomers are all well and good, but cells are much larger than the molecular scale. How do they do it? The cellular machinery links together these single molecular units into long chains of monomers, called polymers. Polymers make up the bulk of what a cell is, dictating its structure and function. Proteins are polymers of amino acids and they drive the cell's chemical reactions or serve as parts of important structures. Starch is a polymer of glucose, and polysaccharides are polymers of sugars that serve in the cell wall, the cell membrane, and as storage products. DNA is a polymer of nucleic acids and is the library that stores the recipes for making an organism.
For polymers made up of more than one monomer, there are several important implications. How each polymer behaves is dictated by the order in which the monomers appear in the polymer sequence. Each specific combination of monomers will create a unique polymer that has a specific structure and function. Changing any one of the monomers in the polymer might change the properties of the polymer. These ideas are especially true for proteins.
A major point here is the economy that this affords to a cell. Instead of needing the ability to fashion several large custom-designed molecules to carry out services within the cell, only a relatively small set of monomers needs to be made. The cell then assembles these to form polymers. This significant efficiency makes organisms very scalable, to borrow a computing term, allowing the construction of microbes only a millionth of a meter in size to blue whales measuring more than 24.5 meters. We will now look at the structure of important monomers in the cell, and the polymers that are made from them.
Sugars are common in the cell
Sugars serve three basic purposes in the cell: as carbon and energy sources, as reservoirs of carbon and energy, and as parts of cellular structures. Large amounts of energy can be extracted from sugars by processes referred to as catabolism, as discussed in the metabolism chapter. This high energy content may explain why many microorganisms prefer sugars if given a choice of energy sources. The term carbohydrate is often used to refer to sugars because their chemical formula can be broken down into [C(H2 O)] x where x is any number greater than three.
Sugar Monomers
Single sugar molecules are typically 3 to 7 carbons long and are termed monosaccharides. Figure 2.13 shows that each carbon on the sugar molecule is decorated with a hydroxyl group (OH) except for one carbon that forms a carbonyl group (C=O). If the carbonyl group is at the end of the molecule, it forms an aldehyde; if the carbonyl group is in the middle of the molecule, it forms a ketone. All sugars can exist as linear molecules in solution. Those greater than five carbons long can also circularize with the carbonyl group attacking a hydroxyl on one of the other carbons. The circular sugar contains 5 to 7 members in the ring, with one of the members being oxygen. Glucose, fructose, and ribose are some of the more common sugars found in the cell.
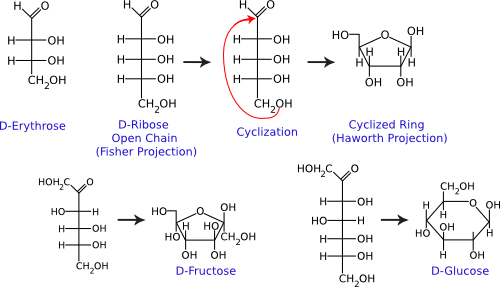
Figure 2.13. Common monosaccharides. The structures of some important monosaccharides are shown. These often serve as building blocks for major structures in the cell, such as the cell wall and the genome. Both the linear and circular forms are shown.
Sugar Polymers
Sugars readily polymerize: the combination of two sugars is called a disaccharide, while the combination of three is called a trisaccharide, and polymers of greater than three sugars are referred to as polysaccharides. Sugars are connected by α or β linkages. If the hydrogen is pointing up, it is an α linkage, and if the hydrogen is pointing down, it is a β linkage, as shown in Figure 2.14. This distinction may seem trivial, but it greatly influences the properties of the molecule. For example, starch is a polymer of glucose linked by α-1,4 bonds. Starch is also water-soluble and can serve as a food source for many organisms. In contrast, cellulose, also containing glucose but linked by β-1,4 bonds, is insoluble in water and is not as readily degraded by most microbes. Polysaccharides might contain only one type of sugar monomer or many, sometimes in repeating units.

Figure 2.14. Common disaccharides, trisaccharides and polysaccharides. Sugars can be linked together to form more complex polymers. Lactose is a common sugar in milk, while maltose is found in many grains. Starch found in potatoes and other vegetables is a long polymer of glucose units. A common form of starch contains 20% amylose and 80% amylopectin.
Polymers of sugars can serve as storage products for the cell. The breakdown of sugar can yield a huge amount of energy, which means that they are terrific molecules for effectively storing energy for later utilization. Starch and glycogen are two examples of sugar polymers that store energy. Polysaccharides also serve as structural components of many different molecules in the cell, including nucleic acids and the cell wall.
Nucleic acids store information and process it
Deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) are involved in information storage and processing. DNA serves as the cell's hereditary information, while RNA converts that information into functional products, such as proteins. Nucleic acids are long polymers of only four nucleotides : adenine, guanine, cytosine, and thymine or uracil. Thymine is in DNA, while uracil is in RNA. Figure 2.15 lists the structure of the five nucleotides found in nucleic acids.
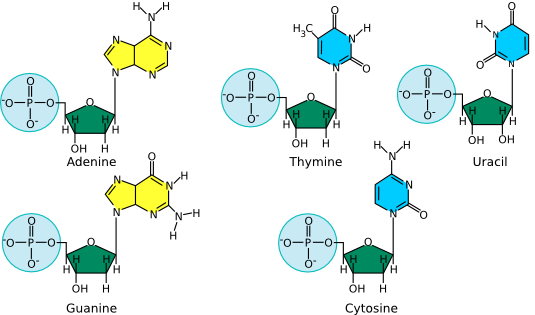
Figure 2.15. The structure of nucleotides. The nucleotides for adenosine (A), guanine (G), cytosine (C ) and thymine (T) as found in DNA are shown. The first three are also found in RNA, but when incorporated into that polymer, the associated sugar has two hydroxyls, as shown in the model of uracil (U). Uracil is the RNA equivalent of the DNA nucleotide thymine.
The nucleotide structure can be broken down into two parts: the sugar-phosphate backbone and the base. All nucleotides share the sugar-phosphate backbone, while the base distinguishes each type of nucleotide. Linking these monomer units together using a 5'-oxygen on the phosphate and the 3'-hydroxyl group on the sugar forms the nucleotide polymer, as shown in Figure 2.16. Many thousands of DNA nucleotides string together to form genes and chromosomes.
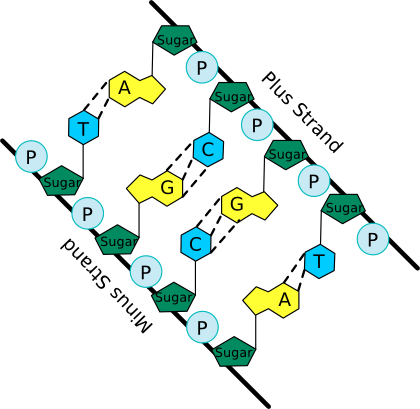
Figure 2.16. A schematic of the nucleic acid polymer. In this picture the bases of the two anti-parallel (having their 5' to 3' linkages running in opposite direction) strands of a DNA double helix are shown. Note the sugar-phosphate backbone from which the bases extend and pair with matching bases from the other strand. The dashed lines show hydrogen bonds between the base pairs.
The bases of the four nucleotides are different, but there is also a pattern. Adenine (A) and guanine (G) are purines, and therefore have a distinctive two-ring structure; they differ in the chemical groups attached to the rings. Likewise, cytosine (C), thymine (T) and uracil (U) are all pyrimidines and share a single-ringed structure, but also differ in their attached groups. Not surprisingly, as these extra chemical groups distinguish the different purines and pyrimidines structurally, they are also responsible for their important functional differences.
The bases in a nucleic acid polymer are capable of forming hydrogen bonds with neighboring bases on a second strand of nucleic acid, a process termed base pairing. However, there are rules to this association. Adenine can form two hydrogen bonds with thymine (or uracil), and cytosine can base pair with guanine, forming three hydrogen bonds. Some of these bonds require the extra chemical groups mentioned above. Suppose two single strands of nucleic acid have sequences that can base pair along the polymer (such sequences are sometimes said to complement). In that case, they will generate a long double-stranded polymer with a staircase topology, as shown in Figure 2.17. This structure is termed a double helix since two strands form the molecule, and it spirals around an axis in a regular pattern. Such a reaction between two complementary DNA strands is spontaneous. If you mix two complementary single strands of nucleic acid in a test tube at a reasonable temperature, pH and salt concentration, they will find each other and anneal to form a double-stranded polymer.
Figure 2.17. The Double Helix. Note how the two nucleic acid strands spiral around each other in a regular repeating pattern. There are 10 bases per turn of the double helix. Bases pair with one another in the center of the helix cylinder and form what some have likened to a spiral staircase. The above figure can be rotated by clicking on the arrows.
Secondary and Tertiary DNA Structures
DNA almost always exists in cells as a double-stranded structure of complementing strands. It happens that this double-stranded form is rather stable. Students often assume that the stability is due to the hydrogen bonding between the bases, but this is not the case (the bases would form hydrogen bonds just as well to water). Instead, it is largely due to the interaction between adjacent base pairs along the helix, which is termed base stacking. Finally, the larger organization of the DNA strands with respect to each other, termed the tertiary structure, is also fairly similar in all DNA molecules. One implication is that proteins that want to distinguish between different DNA molecules must do so by reading different primary structure sequences by interacting with the outer surface of the base pairs, as shown in Figure 2.18.
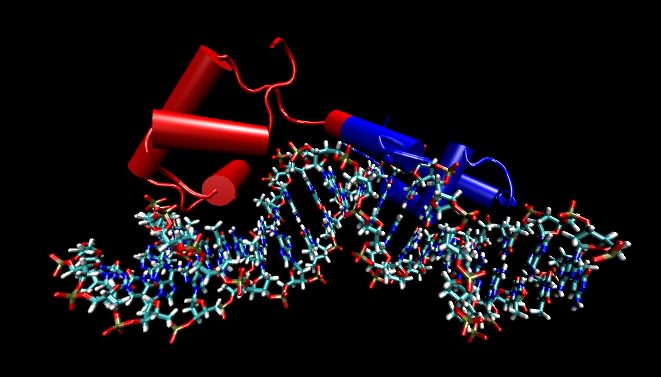
Figure 2.18. A molecular model of the lac repressor. The repressor binding to its recognition sequence. Any DNA-binding protein that recognize specific DNA sequences must make specific contacts with that recognition sequence by reading the pattern of bases in the DNA. These proteins do this by making very specific contacts between themselves and those atoms on DNA that are different in the different bases. In this figure, the DNA helix is shown by the ball-and-stick figure running along the bottom and the protein is depicted by cylinders (which refer to alpha helices) and other "squiggles" (which refer to any other structure in the protein). The point of the figure are that the DNA is not opened up and that the recognition regions of the proteins are very often α helices that lay parallel with or perpendicular to the DNA helix. Not shown are the side chains of the amino acids that actual make the specific contacts with the DNA. In this and subsequent molecular figures, of three different displays will be shown: ball and stick, which shows all atoms larger than hydrogen; ribbon models, in which a ribbon that runs along the carbon backbone of the protein is displayed; and space-filling models, where the actual atomic surface of the molecule is shown. Sometimes, as in this figure, the ribbon form is modified slightly by showing cylinders for alpha helices instead of an actual helix. Most of the structures shown in this text are the product of X-ray crystallography, which essentially displays the position of every atom (larger than hydrogen) in a molecule. Increasingly all biological questions are being thought about in terms of these structures at the atomic scale.
Structures of RNA
In composition and therefore primary structure, RNA is similar to DNA, except that uracil (U) takes the place of thymine in the molecule, and the ribose unit on each sugar contains an additional hydroxyl group. However, most RNA in cells exists as single-stranded molecules and not a complex of two different strands as with DNA. Now, if complementary base sequences are present in an RNA molecule, it can fold back upon itself and base pair so that many RNA molecules have at least some double-stranded regions. However, this bending and folding means that RNA molecules typically have much more complicated tertiary structures than DNA. Both the single-stranded loops and the double-stranded stems are critical for the function of most RNA molecules. Many are involved in creating physical structures, such as ribosomes, that are involved in processing information. The other general class of RNAs are messenger RNAs, which represent a version of the DNA primary structure suitable for translation into protein.
Key Takeaways
- Sugars store carbon and energy and are part of cellular structures.
- Sugars in biological systems are 3 to 7 carbons and contain a carbonyl group.
- Sugars can be monomers or polymers of 2, 3 or more sugars.
- DNA and RNA are composed of two parts, the sugar phosphate backbone and nucleotide bases.
- The bases in DNA are purines (adenine and guanine) and pyrimidines (cytosine and thymine). RNA contains the same bases, except uracil is substituted for thymine and they contain an extra hydroxyl at the 2' carbon.
- DNA polymers can be thousands to millions of base pairs long. RNA polymers are hundreds to thousands of bases long.
- RNA is single-stranded. DNA is double-stranded and forms a double helix in the cell.
- RNA molecules have significant secondary and tertiary structure that is important in their functions.
2 - 6 Proteins are made of amino acids
Learning Objectives
After reading this section, students will be able to...
- Describe the basic structure of an amino acid.
- Explain the peptide bond and how proteins are built.
- Identify the secondary structures that proteins fold into and understand the role of protein motifs in enzyme function.
- Describe tertiary and quaternary structure of proteins.
Proteins and peptides (small proteins) are essential to the cell and serve two major functions. Many proteins are enzymes that catalyze almost all biological reactions in a living organism. Other proteins perform a structural role for the cell - either in the cell wall, the cell membrane or in the cytoplasm. In this section, we will look at the basic structural elements shared by all proteins.
Structure of Amino Acids
Proteins are polymers of amino acids. Amino acids, with rare exception, contain an α carbon that is connected to an amino (NH3 ) group, a carboxyl group (COOH), and a variable side group (R). Figure 2.19 shows this generic amino acid structure. The side group gives each amino acid its distinctive properties and helps dictate the protein's folding.
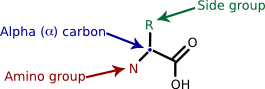
Figure 2.19. A generalized amino acid. Each amino acid contains a carboxyl group, an amino group and a variable side group (R). These all connect to a central carbon, termed the α-carbon, identified by the arrow.
The primary structure of proteins is the amino acid sequence
As with nucleic acids, primary structure refers to the ordered sequence of the different amino acids in a protein. The carboxyl group and the amino group of amino acids are reactive. As shown in Figure 2.20, cells synthesize proteins by attaching the carboxyl group of one amino acid to the amine group of the next, with polymerization taking place at the ribosome. These covalent bonds in proteins are termed peptide bonds. Since each amino acid has a carboxyl group and an amino group, hundreds or thousands of amino acids can be linked together.
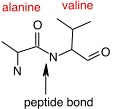
Figure 2.20. A peptide bond. The peptide bond between an alanine residue and a valine residue is identified by the arrow. Peptide bonds can form between any two of the 20 amino acids and link the carboxyl group of one amino acid and the amine group of the next.
There are 20 common amino acids found in proteins, and these amino acids can be roughly classified into three groups: polar, non-polar, and charged. Polar and charged amino acids are hydrophilic and are often found on the surface of a protein, interacting with the surrounding water. In contrast, non-polar (or hydrophobic) amino acids avoid water. While this categorization is adequate for most purposes, you should recognize that it is a bit simplistic. For example, arginine does have a charged hydrophilic group at one end, but the -CH2 - backbone that makes up most of the amino acid is quite hydrophobic. Figure 2.21 shows the chemical structure of all 20 common amino acids.
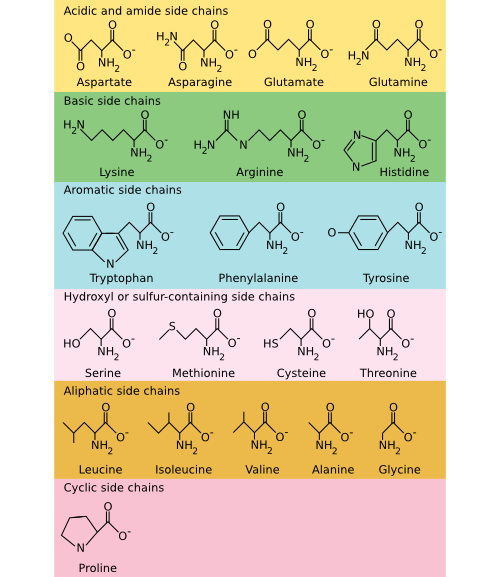
Figure 2.21. The common amino acids. The amino acids are organized into their chemical type and characteristics. Acidic and basic amino acids carry a charge, amino acids with a sulfur or hydroxyl component are polar, and aliphatic and aromatic amino acids are non-polar.
Secondary structure is the local geometry of the protein
Ribosomes, and the rest of the translation machinery, link amino acids together to form peptides and proteins into polymers that range from 10 to 10,000 residues in length. During and after protein synthesis, the residues of the primary sequence dictate how the protein folds. The simplest aspect of protein folding is termed its secondary structure, which refers to the geometry of the local polypeptide chain with respect to its immediate neighbors. While the primary sequence of amino acids dictates protein folding, predicting the overall structure from the primary sequence remains one of biology's most important unsolved problems. Nevertheless, the major determinants of this final structure are hydrophobic interactions. The protein must hide hydrophobic amino acids from the water interface during protein folding by burying them in the interior. This burying defines the protein core, which then influences the immediate structure around it and greatly affects the protein's overall structure.
Common Secondary Structures
Peptide bonds between adjacent amino acids can rotate and twist to allow a large number of interactions. Still, two local organization schemes, the α helix and the β sheet, are found in many proteins. Their prevalence is prevalent because they happen to form particularly energetically favorable structures. Numerous hydrogen bonding opportunities and hydrophobic interactions between nearby amino acids in the protein drive the formation of these structures. The α helix resembles a ribbon of adjacent amino acids wrapped around a tube to form a staircase-like structure. Figure 2.22 shows different representations of an α helix. This structure is very stable, yet flexible and is often present in parts of a protein that may need to bend or move.
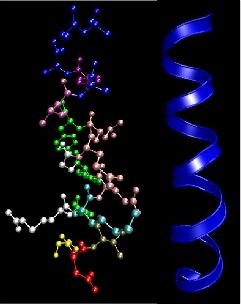
Figure 2.22. The α helix motif in proteins. This figure shows two common depictions of an α-helix, an extremely common and important motif in proteins. The ball-and-stick depiction on the left shows the various side chains, but the helical nature of the structure is not so obvious. The ribbon representation on the right only traces the backbone of the peptide bonds and overemphasizes the symmetry a bit, but shows the helical nature of the structure.
In the β sheet, two protein chains (perhaps different segments of the same protein) align themselves in a planar structure such that hydrogen bonds can form between facing amino acids in each sheet. Figure 2.23 shows different representations of this structure. The β sheet is different from the α helix in that it can involve amino acids from different sections of the protein, which come together to form this structure. Also, the structure tends to be rigid and less flexible than the α helix.
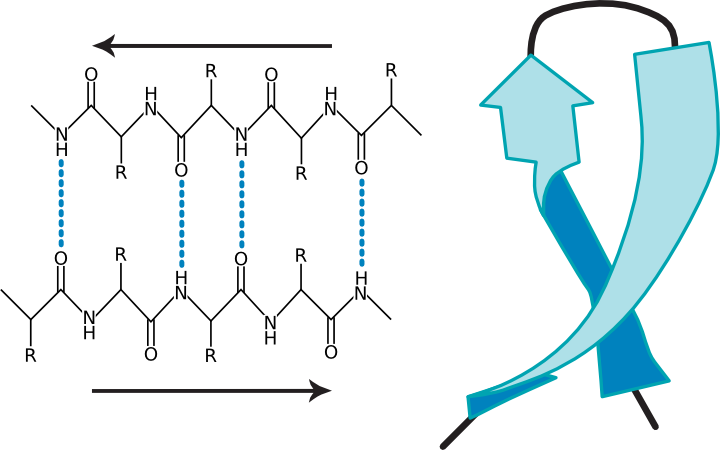
Figure 2.23. Two views of a β-sheet. (a) A diagram of a β sheet showing hydrogen bonding between protein strands (b) A ribbon representation of a β-sheet.
Tertiary structure is the 3D structure of individual polypeptides
The entire protein chain will fold into a series of α helices and β sheets connected by strings of amino acids that form other structures. The overall structure of the folded protein is termed its tertiary structure.
As noted before, the most important stabilizing force in proteins is burying hydrophobic residues from the surrounding water. However, other chemical features are important in creating and stabilizing tertiary structures. These include hydrogen bonds, ionic interactions, and sulfhydryl bonds. Section 2-1 described hydrogen bonds and ionic interactions. Sulfhydryl linkages are unique to proteins and are covalent bonds between cysteine groups. Cysteine is a unique amino acid in that it has a sulfur group at the end of its variable side group that is available for binding to other groups. Often in proteins, nearby sulfhydryl groups on cysteines form a covalent bond, and these are often crucial for the stabilization of the mature protein for it to perform its function. Figure 2.24 shows some representations of sulfhydryl bonds in proteins.
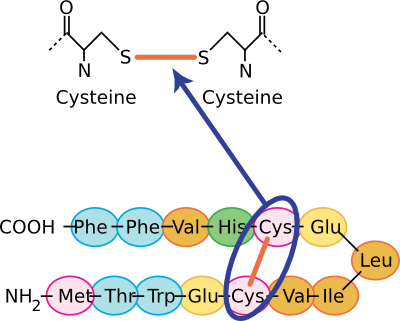
Figure 2.24. The chemical structure of a sulfhydryl bond. Several views of sulfhydryl linkages are shown. In the top panel, the covalent sulfhydryl linkage between two cysteine residues is shown. In the bottom panel, this sort of Cys-Cys bond is shown in the context of a section of a protein.
All of these forces combine to fold a protein into its final tertiary structure. Figure 2.25 shows the complete structure of ribulose bisphosphate carboxylase (RubisCo), one of the most abundant enzymes on this planet. This enzyme takes energy, obtained most often from the sun, and uses it to convert carbon dioxide into carbohydrate. Its abundance reflects the fact that virtually all photosynthetic organisms, from bacteria to plants, use this enzyme to produce much of the cell's carbon. Note the α helices throughout the protein and the β sheet near the bottom. These structures help to hold the protein in its proper conformation so that it can carry out its enzymatic activity.
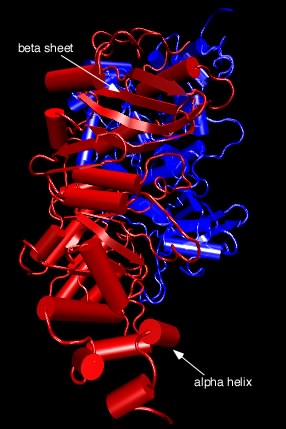
Figure 2.25. Tertiary structure of ribulose bisphosphate carboxylase.. The figure shows the structure of ribulose bisphosphate carboxylase, typically abbreviated Rubisco, from the bacterium Rhodospirillum rubrum . As described in the text, this complex structure has alpha helices (shown as cylinders) and beta sheets (shown as the flat ribbons) throughout.
Now, we have talked about certain secondary structures that recur in many proteins, but it also happens that there are conserved tertiary structures as well. In other words, there are regions of proteins, called motifs or domains, that are structurally identical in two or more different proteins. Figure 2.26 shows some examples of protein motifs. By "structurally identical," we mean that they have similarly sized α helices and β sheets organized in a similar overall way. Interestingly, these domains do not always have identical primary structures. Instead, it appears that a variety of different sequences can fold to give the same overall tertiary form. However, they will often have conserved amino acids in them at critical positions. These motifs often have a specific function. For example, there is a widespread motif that proteins use for binding ATP. A second class of examples would be the set of motifs that help bind DNA. Through genetic changes, these motifs have been grafted onto other protein regions so that the same ATP-binding (or the DNA-binding) domain performs a similar function for proteins with completely different overall activities. Of course, the unique functions are defined by those portions of the protein that are not in common.
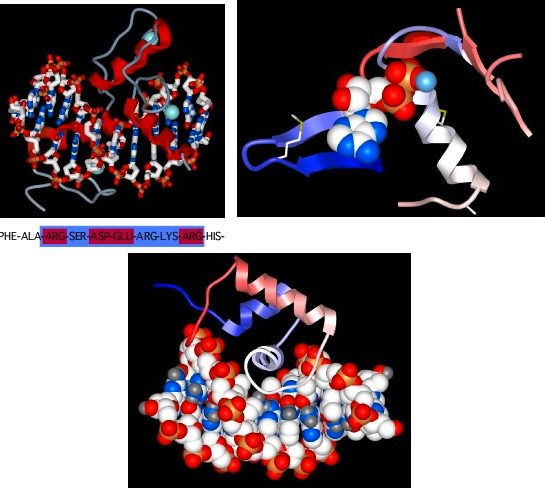
Figure 2.26. Some common protein motifs. Three examples of protein motifs are shown. The top left panel shows two zinc-finger motifs (the red helices and thin gray lines), each bound to a Zinc atom shown in pale blue. This is a common motif found in DNA-binding proteins and the interaction is shown here (the ball-and-stick figure running from left to right in this panel is a section of DNA). Note that the Zinc does not interact with the DNA, but instead stabilizes the protein motif to allow it to form the precise shape to bind to DNA. The top right panel shows an ATP-binding domain, with the ATP displayed as the space-filling model and the protein shown in a ribbon format. The bottom panel shows another common DNA-binding motif, termed the helix-turn-helix. Here the DNA is shown in space-filling form and the two helices after which the structure is named are shown in red and white (the latter is shown end-on and is interacting with the DNA).
From this description, you think of proteins as a collection of protein pieces, each of which serves a function and has an evolutionary history. We refer to these as functional domains. The enzyme pyruvate kinase, which generates ATP by removing it from phosphoenolpyruvate, has three domains, termed A, B, and C. The A domain is responsible for binding substrate (phosphoenolpyruvate and ADP) while the C domain serves a regulatory role, binding fructose-6-bisphosphate and stimulating activity.
Up to this point, we have talked as if proteins are static, rigid structures, but this is certainly wrong. Instead, most proteins are flexible, dynamic structures that respond to the conditions around them. Movement is typically essential to their function, especially for enzymes that need to grab a substrate, react with it, and release product. This movement can be just a few tenths of an angstrom (10 -10 meters), while in other proteins, this movement can be quite large. This shift obviously changes the tertiary structure of that protein. Thus, when we talk about tertiary structures, typically determined by crystallography, we refer to only one of the possible structures that the protein can exist in while performing its biological function.
Quaternary structure is the total complex of a functional protein
Many proteins are actually complexes of several polypeptides. The quaternary structure of a protein is the arrangement of more than a single polypeptide into a collection. Protein complexes might contain two or more copies of the same protein, or they may consist of any number of different polypeptides in various ratios. Such complexes are certainly not random but reflect precise interactions among the protein subunits based on the same sorts of interactions (e.g., hydrophobic, hydrogen bonds, etc.) described above.
Figure 2.27 shows the catabolite activator protein, an example of a protein containing identical subunits. In this case, it has two subunits, termed a dimer. Each subunit has a cyclic adenosine monophosphate molecule bound that activates the protein. In the active state, this protein binds DNA and activates various genes in the cell.
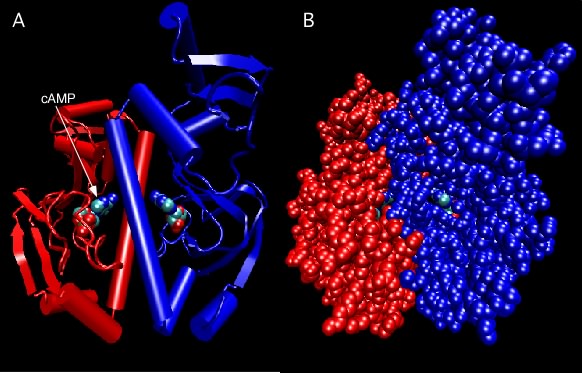
Figure 2.27. Catabolite Activator Protein structure. Two depictions of the catabolite activator protein (CAP) of E. coli are shown. The left panel shows a ribbon depiction, with the two identical subunits shown in red and blue. CAP senses the presence of cyclic AMP (cAMP) in the cell and a molecule of cAMP bound to each monomer as indicated. The dimer is actually symmetrical, but that is not obvious here because the model is rotated slightly. The right panel shows a ball-and-stick representation of the same dimer and gives a better sense of the overall shape. Note that the bound cAMP molecules are buried within the protein (so how do they ever come and go?).
Dinitrogenase is an example of a protein containing non-identical subunits. As shown in Figure 2.28, the active protein has two copies of one protein (termed α) and two copies of a second protein (termed β), and therefore is also referred to as an α 2 β 2 tetramer. This protein is responsible for the reduction of N2 gas to ammonia and is critical to global nitrogen cycling. The importance of the quaternary structure of dinitrogenase is only partially clear. The α subunits contain the metal cluster where N2 is reduced (or "fixed") and the β subunit helps form another metal center that helps transfer electrons to that active site. We can therefore understand why there might need to be an αβ dimer. However, it is unclear why there are two such αβ dimers hooked together in the tetramer found in nature. There is, for example, no apparent communication between the two active sites.
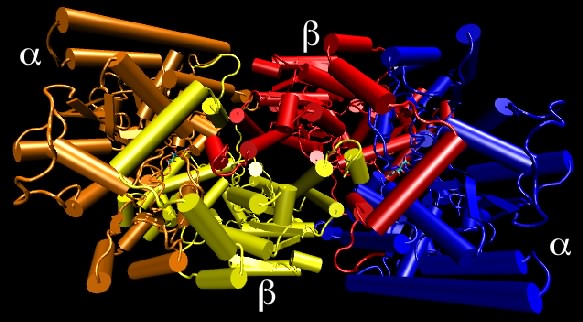
Figure 2.28. Nitrogenase structure. Nitrogenase is made up of four protein chains, with two copies each of different proteins, termed the α and β subunits. Though not apparent in this view, the two α subunits are identical to each other, as are the two β subunits. The quaternary structure is referred to as an α2β2 tetramer.
Glycoproteins are proteins with attached sugars that are typically important for the proper function of the protein. In most cases, enzymes add the sugars to the protein after translation. Proteins that encounter the outside environment of the cell are sometimes glycosylated to stabilize them against attack from degradative enzymes and destructive physical forces.
Key Takeaways
- Amino acids are organic acids with an amine group attached to the α carbon. Amino acids are distinguished by their side group that dictates their properties.
- Proteins contain 20 different amino acids that are linked by peptide bonds to form long polymers. Amino acids can be classified into three groups, polar, non-polar, and charged.
- The chain of amino acids dictates how a protein folds. The major determinant of protein folding is hydrophobic interactions. The local geometry of a protein is its secondary structure. Two common secondary structures are the α helix and the β sheet.
- Tertiary structure is stabilized by hydrophobic interactions, hydrogen bonds, ionic interactions, and sulfhydryl bonds.
- Conserved tertiary structures are called motifs. Many common motifs have been discovered. Some examples are ATP-binding and DNA-binding motifs.
- Proteins are flexible, dynamic structures that respond to their surroundings.
2 - 7 Lipids and small molecules
Learning Objectives
After reading this section, students will be able to...
- Describe the basic structure of a lipid.
- Identify the small molecules that are important for cellular function and their roles.
Lipids are molecules with two personalities. One part of the molecule wants to associate with water, and the other does not. Molecules with these properties are termed amphipathic. Figure 2.29 shows that the backbone of the lipid consists of a three-carbon glycerol molecule. Hydrophobic, long-chain fatty acids attach to two hydroxyl groups on the glycerol. To the third hydroxyl group, a polar, and therefore hydrophilic, group is attached. Many bacteria contain phospholipids in which this third group contains a phosphate connected to a carbon molecule. The amphipathic nature of lipids is important in their function in the cell.
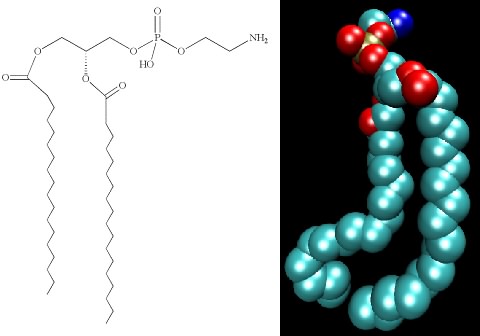
Figure 2.29. The structure of phosphatidylethanolamine. The chemical structure (left) and a space-filling model (right) of phosphatidylethanolamine.
Most lipids are phospholipids, but about 50% of all known bacterial DNA also contain hopanoids, as shown in Figure 2.30. These molecules have a similar structure to sterols found in eukaryotic membranes and serve to help stabilize the membrane. Lipids will self assemble and form the cellular membrane, and Section 3-2 describes this process in greater detail.
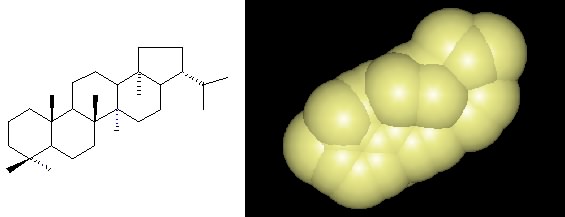
Figure 2.30. A hopanoid. The chemical structure and space-filling model of a hopanoid, which is found in many different bacterial membranes.
Small molecules are also important in the cell
Several essential small molecules shuttle protons, electrons, or small carbon moieties around the cell. These small entities typically do their job in association with proteins to which they can be either loosely or tightly bound. All life on this planet seems to have settled on a surprisingly small set of molecules to perform these tasks. Almost certainly, the use of these molecules evolved early and has remained the same through the ages.
Proton and electron carriers
Most amino acids are not particularly good at either donating or accepting electrons and when they do, it is under a limited range of conditions. As you will read in the chapter on metabolism, the ability to move electrons among proteins is critical to all life, so two general types of prosthetic groups associated with proteins have evolved for this purpose. Figure 2.31 shows the two types of structures commonly shuttle electrons: organic, multi-ring structures and iron-sulfur clusters. In both cases, these carriers have characteristic affinities for accepting and donating electrons and protons. However, the surrounding proteins modulate these affinities. Thus, a wide range of electron carriers with different properties has evolved. By organizing these carriers in precise patterns in the cell, the cell can use the transfer of electrons to do work.
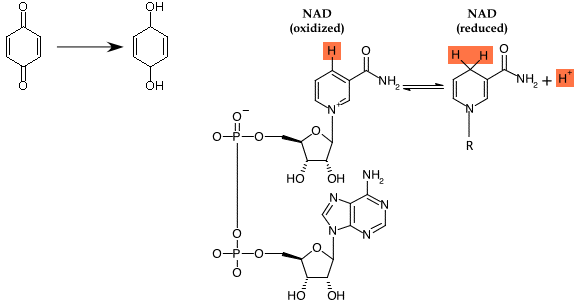
Figure 2.31. The structures of a few important electron and hydrogen carriers. The chemical structures of quinone (a) and nicotinamide adenine dinucleotide (NAD) (b) are both organic electrons carriers. The cell also has the need for inorganic electron carriers, most often Fe. The structure of an Fe-S center is shown in (c). In each case, both structures the oxidized and reduced forms are depicted.
Carbon carriers
Small molecules in the cell also serve as carriers of important carbon compounds. Essentially, these carriers have the right chemical properties that make it relatively easy for enzymes to add or remove a particular carbon unit. Tetrahydrofolate and cobalamin (vitamin B12 ) are often involved in adding or removing one-carbon units during the synthesis of various structures in the cell. Coenzyme A is necessary for the transfer of small 2 to 4 carbon units (acetyl, propyl) from one enzyme to another. It finds utility in both the synthesis and breakdown of organic molecules. The beauty of using a small set of carriers is that it allows the easy movement of carbon from one pathway to another.
Important minerals
Many types of minerals are important for the proper functioning of enzymes. For example, magnesium ions are essential for ATP-binding by many enzymes. Zinc is important in the proper folding of some enzymes and iron, in the form of iron-sulfur centers and hemes, is critical in many electron transport proteins. Minerals also help bind structures in the cell together. For example, magnesium and calcium are necessary for the stabilization of membranes. Potassium ions in the cell shield the large amount of negative charge on the DNA allowing it to pack more tightly together. More will be said in later chapters about their specific roles, but some of the more important ions include K + , PO4 -3 , Mg +2 , Zn +2 , Ca +2 , Mn +2 , Fe +2 and Fe +3 .
Key Takeaways
- Membranes have lipids as their major constituent.
- Lipids contain a glycerol backbone. To this backbone are attached a polar group and two long-chain fatty acids.
- NAD and FAD are two common proton and electron carriers in the cell. Inorganic Fe-S centers are also important electrons carriers.
- Tetrahydrofolate, cobalamin and coenzyme A are common carbon carriers in the cell.
- Many enzymes require minerals for proper function in the cell. Common examples include, iron, zinc, magnesium, and calcium.
2 - 8 Summary
Everything we can see, touch, taste, or experience in any way consists of matter. Matter is atoms, and they consist of the subatomic particles, protons, neutrons, and electrons. Biological compounds are matter, just like everything else, but living systems tend to be made of the lighter elements, with C, N, O, H, P, and S being the majority. These elements are part of cell structures, and many of these structures are formed by making polymers. These polymers consist of just a few basic types of molecules: amino acids, sugars, nucleic acids, and fatty acids. Sugars are used in structures in the cell, and sometimes decorate proteins and lipids. Nucleic acids are more complex polymers having a backbone made of sugar and phosphorus, with the base giving the nucleic acid its identity. Proteins are polymers of 20 assorted amino acids, and they are the biological catalysts of the cell. The sequence of the polymers dictates their structure and subsequently their function, especially for proteins. Finally, lipids are made from glycerol, a polar head group, and two long-chain fatty acids. The amphipathic nature of lipids encourages them to assemble into a lipid bilayer and forms the foundation of the cell. In subsequent chapters, we will discuss how this chemistry comes together to form the diversity of living organisms we call microbes.
If you enjoyed this free chapter on the fundamentals of structure, consider buying the book, for only $30.